Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
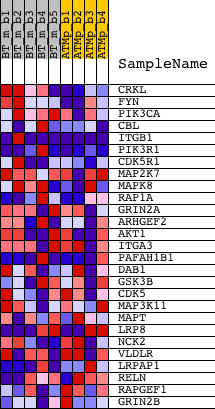
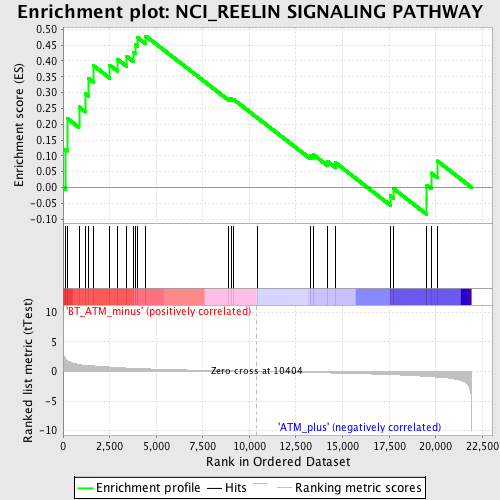
| GeneSet | NCI_REELIN SIGNALING PATHWAY |
| Enrichment Score (ES) | 0.47836265 |
| Normalized Enrichment Score (NES) | 1.4961003 |
| Nominal p-value | 0.033707865 |
| FDR q-value | 0.639136 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CRKL | 1421953_at 1421954_at 1425604_at 1436950_at | 122 | 2.239 | 0.1195 | Yes | ||
| 2 | FYN | 1417558_at 1441647_at 1448765_at | 216 | 1.850 | 0.2186 | Yes | ||
| 3 | PIK3CA | 1423144_at 1429434_at 1429435_x_at 1440054_at 1453134_at 1460326_at | 854 | 1.165 | 0.2545 | Yes | ||
| 4 | CBL | 1434829_at 1446608_at 1450457_at 1455886_at | 1207 | 1.030 | 0.2960 | Yes | ||
| 5 | ITGB1 | 1426918_at 1426919_at 1426920_x_at 1427771_x_at 1438119_at 1452545_a_at | 1377 | 0.995 | 0.3439 | Yes | ||
| 6 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 1613 | 0.946 | 0.3860 | Yes | ||
| 7 | CDK5R1 | 1421123_at 1421124_at 1433450_at 1433451_at | 2504 | 0.741 | 0.3867 | Yes | ||
| 8 | MAP2K7 | 1421416_at 1425393_a_at 1425512_at 1425513_at 1440442_at 1451736_a_at 1457182_at | 2920 | 0.662 | 0.4047 | Yes | ||
| 9 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 3414 | 0.576 | 0.4144 | Yes | ||
| 10 | RAP1A | 1424139_at | 3762 | 0.526 | 0.4279 | Yes | ||
| 11 | GRIN2A | 1421616_at | 3884 | 0.512 | 0.4510 | Yes | ||
| 12 | ARHGEF2 | 1421042_at 1421043_s_at 1427646_a_at | 3986 | 0.501 | 0.4744 | Yes | ||
| 13 | AKT1 | 1416657_at 1425711_a_at 1440950_at 1442759_at | 4445 | 0.446 | 0.4784 | Yes | ||
| 14 | ITGA3 | 1421997_s_at 1455158_at 1460305_at | 8891 | 0.096 | 0.2808 | No | ||
| 15 | PAFAH1B1 | 1417086_at 1427703_at 1439656_at 1441404_at 1444125_at 1448578_at 1456947_at 1460199_a_at | 9016 | 0.089 | 0.2801 | No | ||
| 16 | DAB1 | 1421100_a_at 1427307_a_at 1427308_at 1435577_at 1435578_s_at | 9159 | 0.080 | 0.2781 | No | ||
| 17 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 10461 | -0.003 | 0.2189 | No | ||
| 18 | CDK5 | 1422590_at 1450674_at | 13272 | -0.179 | 0.1006 | No | ||
| 19 | MAP3K11 | 1450669_at | 13441 | -0.190 | 0.1036 | No | ||
| 20 | MAPT | 1417885_at 1424718_at 1424719_a_at 1445634_at 1455028_at | 14182 | -0.240 | 0.0832 | No | ||
| 21 | LRP8 | 1421459_a_at 1440882_at 1442347_at | 14618 | -0.273 | 0.0786 | No | ||
| 22 | NCK2 | 1416796_at 1416797_at 1458432_at | 17570 | -0.528 | -0.0266 | No | ||
| 23 | VLDLR | 1417900_a_at 1434465_x_at 1435893_at 1438258_at 1442169_at 1451156_s_at | 17758 | -0.551 | -0.0044 | No | ||
| 24 | LRPAP1 | 1426696_at 1426697_a_at 1436609_a_at 1452148_at | 19535 | -0.828 | -0.0392 | No | ||
| 25 | RELN | 1449465_at 1458020_at | 19537 | -0.828 | 0.0070 | No | ||
| 26 | RAPGEF1 | 1421146_at 1427006_at 1441386_at | 19760 | -0.875 | 0.0458 | No | ||
| 27 | GRIN2B | 1422223_at 1431700_at | 20100 | -0.957 | 0.0837 | No |